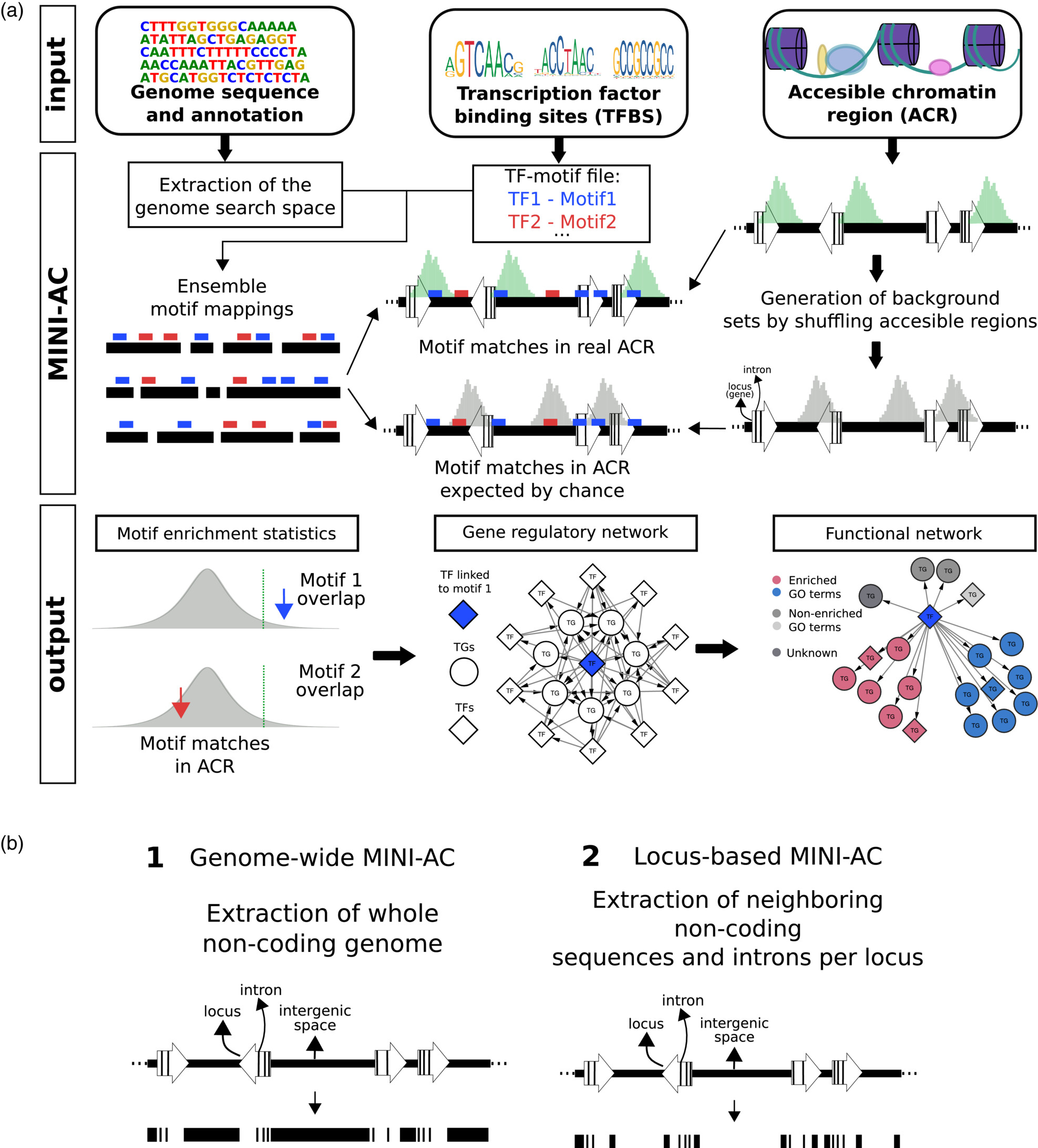
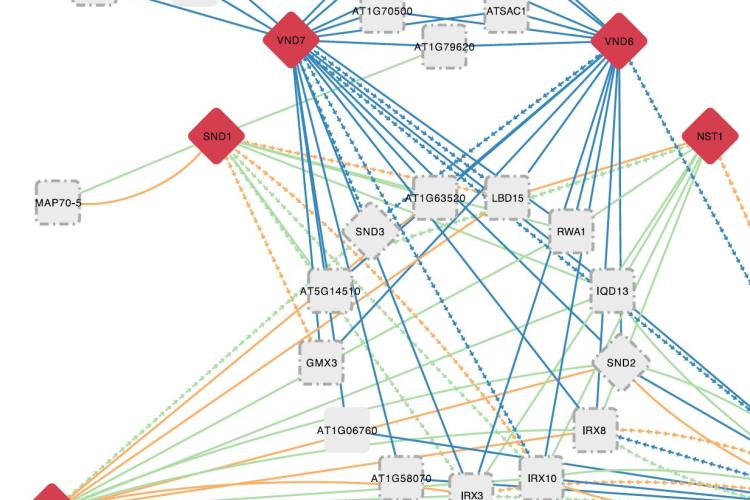
Through the integration of large-scale experimental datasets and the application of computational methods, we start to have functional clues for an increasing number of genes in plants. Although new technological developments improve the resolution of measuring for example when and where a gene is expressed, understanding the underlying mechanisms and identifying the sequences and factors controlling gene expression remains a major challenge. Transcription factors (TFs) are important regulators mediating gene expression and different genomic technologies have recently been applied to start mapping the regulatory interactions between TFs and target genes in plants. As there are approximately 1700 Arabidopsis TFs and thousands of potential target genes, current gene regulatory networks remain incomplete. We develop and apply new computational approaches to identify, based on raw data from ChIP-chip/Seq, ATAC-Seq, protein binding microarrays, TF perturbation data or phylogenetic footprinting, new TF-target interactions and perform systematic regulatory gene annotations. By combining specific bulk and single-cell RNA-Seq datasets with newly inferred regulatory interactions, we develop new approaches to explore condition-specific gene regulatory networks, characterize associated cis-regulatory sequences, and shed light on the functions of TFs in different biological contexts (e.g., De Clercq et al., 2021; MINI-EX case studies). Through sequence-, expression-, and chromatin-based methods, we also characterize regulatory sequences and networks in crop species (e.g., maize ; MINI-AC). More recently, different machine learning models are used to unveil cis-regulatory sequences controlling context-specific expression (Smet et al., 2023).
Selected references:
- MINI-AC: inference of plant gene regulatory networks using bulk or single-cell accessible chromatin profiles. Manosalva Pérez N, Ferrari C, Engelhorn J, Depuydt T, Nelissen H, Hartwig T, Vandepoele K. Plant Journal. 2024 Jan;117(1):280-301. doi: 10.1111/tpj.16483
- Transcription factors KANADI 1, MYB DOMAIN PROTEIN 44, and PHYTOCHROME INTERACTING FACTOR 4 regulate long intergenic noncoding RNAs expressed in Arabidopsis roots. Liu L, Heidecker M, Depuydt T, Manosalva Perez N, Crespi M, Blein T, Vandepoele K. Plant Physiol. 2023 Oct 26;193(3):1933-1953. doi: 10.1093/plphys/kiad360
- MINI-EX: Integrative inference of single-cell gene regulatory networks in plants. Ferrari C, Manosalva Pérez N, Vandepoele K. Molecular Plant. 2022 Nov 7;15(11):1807-1824. doi: 10.1016/j.molp.2022.10.016
- Integrative inference of transcriptional networks in Arabidopsis yields novel ROS signalling regulators. De Clercq I, Van de Velde J, Luo X, Liu L, Storme V, Van Bel M, Pottie R, Vaneechoutte D, Van Breusegem F, Vandepoele K. Nature Plants. 2021 Apr;7(4):500-513. doi: 10.1038/s41477-021-00894-1
- Predicting transcriptional responses to heat and drought stress from genomic features using a machine learning approach in rice. Smet D, Opdebeeck H, Vandepoele K. Front Plant Sci. 2023 Jul 17;14:1212073. doi: 10.3389/fpls.2023.1212073