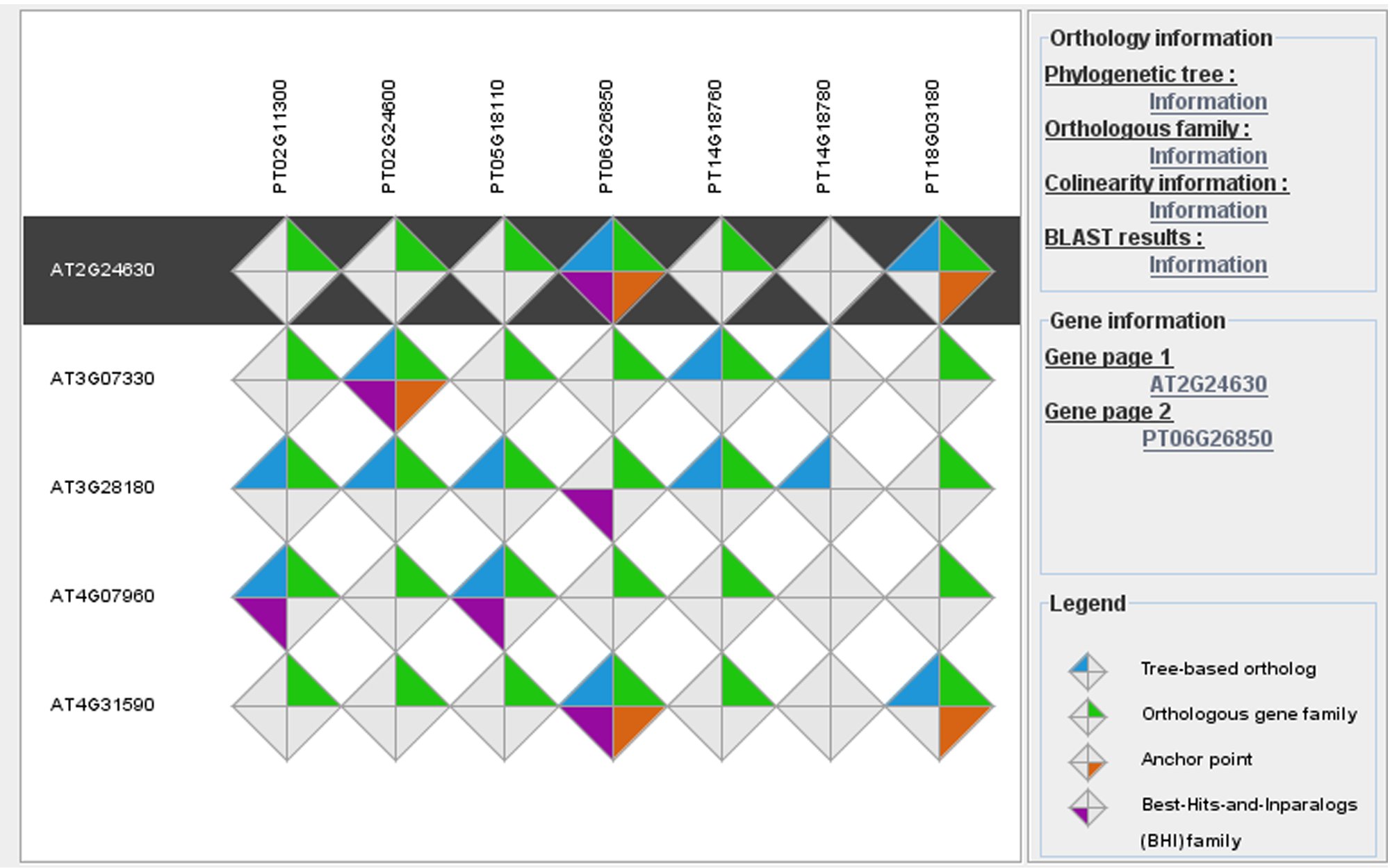
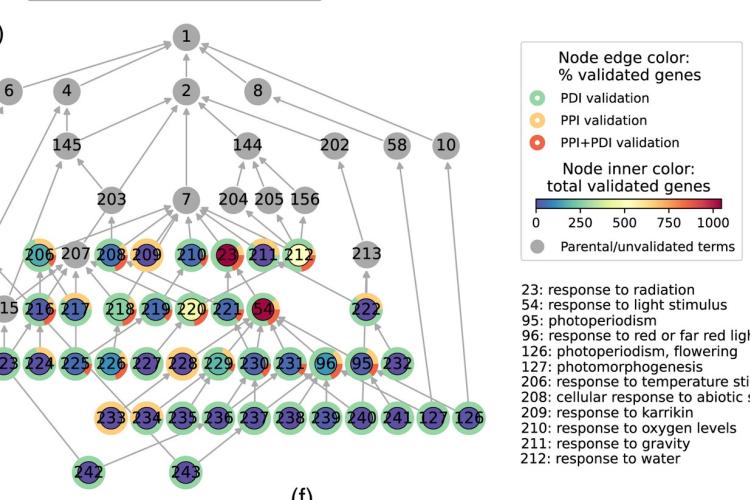
Plant genomes contain an extremely large number of genes, both in number of gene families as well as in copy number per family. Consequently, searching for individual genes with conserved biological functions across different plant species is challenging. Apart from exploring known gene function information (e.g., Gene Ontology, literature/text mining), when and where a gene is expressed is an important factor contributing to the concept of gene function in multicellular organisms. Therefore, comparing expression data between different species offers a valuable alternative approach to identify genes showing conserved expression profiles and, as such, to translate detailed biological knowledge from model species to crops (e.g., Curci et al., 2022). Apart from comparative sequence analysis tools (e.g., PLAZA), we develop new methods to integrate experimental information through multi-omics network approaches and to identify new genes and functional modules involved in specific biological processes (e.g., De Clercq et al., 2021 ; Depuydt et al., 2021, vHRR), focusing both on protein-coding and lincRNA genes (e.g., Li et al., 2023).
Selected references:
- Application of orthology and network biology to infer gene functions in non-model plants. Vandepoele K, Thierens S, Van Bel M. Physiol Plant. 2024 Jul-Aug;176(4):e14441. https://doi.org/10.1111/ppl.14441 #Review
- Transcription factors KANADI 1, MYB DOMAIN PROTEIN 44, and PHYTOCHROME INTERACTING FACTOR 4 regulate long intergenic noncoding RNAs expressed in Arabidopsis roots. Liu L, Heidecker M, Depuydt T, Manosalva Perez N, Crespi M, Blein T, Vandepoele K. Plant Physiol. 2023 Oct 26;193(3):1933-1953. doi: 10.1093/plphys/kiad360
- Identification of growth regulators using cross-species network analysis in plants. Curci PL, Zhang J, Mähler N, Seyfferth C, Mannapperuma C, Diels T, Van Hautegem T, Jonsen D, Street N, Hvidsten TR, Hertzberg M, Nilsson O, Inzé D, Nelissen H, Vandepoele K. Plant Physiol. 2022 Nov 28;190(4):2350-2365. doi: 10.1093/plphys/kiac374
- Charting plant gene functions in the multi-omics and single-cell era. Depuydt T, De Rybel B, Vandepoele K. Trends Plant Sci. 2023 Mar;28(3):283-296. doi: 10.1016/j.tplants.2022.09.008 #Review
- Integrative inference of transcriptional networks in Arabidopsis yields novel ROS signalling regulators. De Clercq I, Van de Velde J, Luo X, Liu L, Storme V, Van Bel M, Pottie R, Vaneechoutte D, Van Breusegem F, Vandepoele K. Nature Plants. 2021 Apr;7(4):500-513. doi: 10.1038/s41477-021-00894-1
- PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants. Van Bel M, Silvestri F, Weitz EM, Kreft L, Botzki A, Coppens F, Vandepoele K. Nucleic Acids Res. 2022 Jan 7;50(D1):D1468-D1474. doi: 10.1093/nar/gkab1024